The labeling of a transcription factor in this fashion allows for the easy and rapid determination of its approximate molecular weight in a denaturing polyacrylamide gel even in crude extracts. Frequently, halogenated analogues of thymidine for example, bromodeoxyuridine or BrdU are incorporated into the DNA enzymatically to enhance the crosslinking between protein and DNA.
As the name implies, Southwestern blotting is a variation of the Western blotting technique. Cell extracts containing the DNA-binding protein of interest are resolved by denaturing polyacrylamide gel electrophoresis followed by electrophoretic transfer to a nitrocellulose membrane. The membrane is then probed with a radioactively labeled DNA fragment bearing the recognition site, preferably in the form of tandem repeats.
The protein that interacts with the probe can be visualized by autoradiography after nonspecifically bound DNA is first washed away from the membrane. To characterize the biochemical properties of transcription factors, it is often necessary to study them in pure cloned forms. Several different approaches have recently been developed to achieve the cloning of cDNAs encoding various transcription factors. They generally fall into two major categories: A major difficulty in the purification of transcription factors is their low abundance ranging between 10 3 and 10 5 molecules per cell.
Another requirement is that a DNA sequence with high affinity and specificity for the transcription factor should be identified so that the DNA binding activity of the protein can be monitored during each step of the purification process. The biochemical purification of a transcription factor begins with the preparation of nuclear extracts from appropriate cells or tissues. This step gives between and fold enrichment of the nuclear-localizing protein. Ammonium sulfate fractionation is frequently used to further concentrate the protein in the nuclear extracts.
Transcription factors (article) | Khan Academy
Following dialysis to remove the ammonium sulfate, the protein mixtures are then subjected to fractionation by conventional column chromatography. A number of different chromatographic procedures can be employed. Examples include gel filtration chromatography such as Sephacryl S, heparin-agarose affinity chromatography and DNA-cellulose chromatography. The latter two are based on the ability of most transcription factors to bind to negatively charged heparin and to interact with random DNA sequences, respectively.
After washing the loaded column, proteins adsorbed to the column are eluted with appropriate buffers and collected in fractions. The fractions that contain the highest activity are pooled and subjected to the next step of purification, which frequently involves sequence-specific DNA affinity chromatography Kadonaga and Tjian , Rosenfeld and Kelly To construct the column, multiple tandem repeats of a double-stranded oligonucleotide bearing the high affinity-binding sequence for the transcription factor are covalently attached to a matrix support such as cyanogen bromide-activated Sepharose beads.
After passing the proteins through this column, the transcription factor will bind to the matrix, which can then be eluted with a salt gradient. The procedure can be repeated to increase the protein purity. Many transcription factors can be purified up to fold by two sequential DNA affinity chromatographic steps. Two recently described techniques can be used in expression cloning, which is based primarily on the sequence-specific interaction of a transcription factor with its high affinity-binding site. One involves the screening of an expression cDNA library with a radiolabeled oligonucleotide probe containing the recognition site for the transcription factor Singh et al.
The other is a genetic selection using the yeast one-hybrid system Wang and Reed Both will be briefly discussed below. The principal of this technique is similar to that of Southwestern blotting. The induced proteins are transferred to nitrocellulose membranes followed by an optional step of denaturation and stepwise renaturation. The membranes are then probed with radiolabeled tandem repeats of an oligonucleotide bearing the high affinity-binding sequence.
After washing the membranes to remove any nonspecifically attached probes, autoradiography is performed to identify the cDNA clones that bind to the probe. The authenticity of these clones can subsequently be verified by binding crude extracts prepared from lysogens of the recombinant phage to the radiolabeled probe using EMSA or DNase I footprinting assay. The yeast one-hybrid system stems from the important observation that most eukaryotic transcription factors are modular by nature, composed of a target-specific DNA-binding domain and a target-independent transcription activation domain.
In this procedure Fig. A reporter construct is generated that consists of multiple copies of the target sequence X adjacent to a low activity promoter Pmin directing HIS3 gene expression. The further introduction of a GAL4-candidate fusion protein capable of binding to the target sequence present in the reporter will strongly activate transcription and increase levels of HIS3. This allows rapid growth and subsequent positive identification of candidate clones in media that lack histidine.
Cloning of transcription factors by the yeast one-hybrid system. A reporter plasmid is constructed in a yeast vector that contains the HIS3 reporter driven by a minimal promoter Pmin. This vector is linked to three tandem copies of the binding sequence X to which the transcription factor X TF X binds. The stably transformed yeast strain is then used to screen a library containing the AD of GAL4 fused to a library of cDNA sequences obtained from a given cell or tissue.
Transcription factors are often classified based on the structural motifs that constitute their DNA-binding domains. In most cases the protein makes a large number of contacts with the DNA, involving hydrogen bonds, ionic bonds and hydrophobic interactions. Although each individual contact is weak, the 20 or so contacts that typically form at the protein-DNA interface ensure that the interaction is both specific and strong.
In this section several major classes of eukaryotic transcription factors Fig. Readers are reminded that transcription of eukaryotic genes requires the participation of many additional regulatory proteins other than the sequence-specific DNA-binding transcription factors addressed below.
Several recent articles provide excellent reviews of these topics Glass et al. Major classes of eukaryotic transcription factors. The part containing helices 2 and 3 closely resembles the helix-turn-helix motif with the recognition helix helix 3 making important contacts with the major grove B. This protein belongs to the Cys-Cys-His-His C 2 H 2 family of zinc finger proteins, named after the amino acids that grasp the zinc.
The zinc finger transcription factors often contain multiple zinc fingers that are contiguous to each other and contact the DNA in similar ways. In D a small sphere represents the zinc atom in each finger. E A leucine zipper dimer bound to DNA. F A helix-loop-helix dimer bound to DNA. The termination of transcription of pre-rRNA genes by polymerase Pol I is performed by a system that needs a specific transcription termination factor.
Termination of transcription occurs in the ribosomal intergenic spacer region that contains several transcription termination sites upstream of a Pol I pausing site. The polymerase continues to move along the template, generating a second RNA molecule associated with the elongation complex.
Polymerase is released as the highly processive exonuclease overtakes it. It is proposed that an emerging view will express a merge of these two models. RNA polymerase III can terminate transcription efficiently without the involvement of additional factors. The Pol III termination signal consists of a stretch of thymines on the nontemplate strand located within 40bp downstream from the 3' end of mature RNAs.
RNA-duplex-dependent termination is an ancient mechanism that dates back to the last universal common ancestor. The regulation of gene expression in eukaryotes is achieved through the interaction of several levels of control that acts both locally to turn on or off individual genes in response to a specific cellular need and globally to maintain a chromatin-wide gene expression pattern that shapes cell identity.
Transcription requires displacement of the positioned nucleosomes to enable the transcriptional machinery to gain access of the DNA. All steps in the transcription are subject to some degree of regulation. Targeting the rate-limiting initial step is the most efficient in terms of energy costs for the cell. Transcription initiation is regulated by cis-acting elements enhancers, silencers, isolators within the regulatory regions of the DNA, and sequence-specific trans-acting factors that act as activators or repressors. The eukaryotic genome is organized into a compact chromatin structure that allows only regulated access to DNA.
The chromatin structure can be globally "open" and more transcriptionally permissive, or globally "condensed" and transcriptionally inactive. The former euchromatin is lightly packed and rich in genes under active transcription. The latter heterochromatin includes gene-poor regions such as telomeres and centromeres but also regions with normal gene density but transcriptionally silenced. The gene expression patterns that define cell identity are inherited through cell division.
DNA methylation is reliably inherited through the action of maintenance methylases that modify the nascent DNA strand generated by replication.
DNA BINDING ASSAYS USED TO STUDY TRANSCRIPTION FACTORS
Specialized proteins can recognize the marker and recruit histone deacetylases and methylases to re-establish the silencing. Nucleosome histone modifications could also be inherited during cell division, however, it is not clear whether it can work independently without the direction by DNA methylation. The two main tasks of transcription initiation are to provide RNA polymerase with an access to the promoter and to assemble general transcription factors with polymerase into a transcription initiation complex.
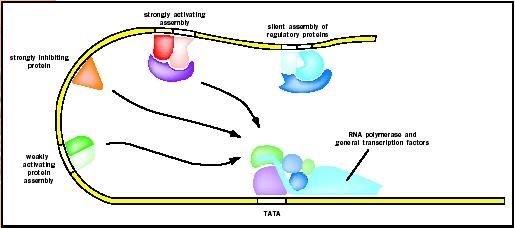
Diverse mechanisms of initiating transcription by overriding inhibitory signals at the gene promoter have been identified. Enhancers can facilitate highly cooperative action of several transcription factors which constitute enhanceosomes. Remote enhancers allow transcription regulation at a distance.
Insulators situated between enhancers and promoters help define the genes that an enhancer can or cannot influence. Eukaryotic transcriptional activators have separate DNA-binding and activating functions. An activator can also recruit nucleosome modifiers that alter chromatin in the vicinity of the promoter and thereby help initiation.
- Eukaryotic transcription factors. - PubMed - NCBI;
- Eukaryotic transcription.
- Eukaryotic Transcription Factors | ScienceDirect.
- Dana und Bianca: Das Abenteuer der Sechser-Bande (German Edition)!
- Transcription factor;
- Eukaryotic transcription - Wikipedia!
- A History of the Earth and Its Mass Extinctions: This Time Its Us.
Multiple activators can work together, either by recruiting a common or two mutually dependent components of the transcriptional machinery, or by helping each other bind to their DNA sites. Eukaryotic transcription repressors share some of the mechanisms used by their prokaryotic counterparts. For example, by binding to a site on DNA that overlaps with the binding site of an activator, a repressor can inhibit binding of the activator. But more frequently, eukaryotic repressors inhibit the function of an activator by masking its activating domain, preventing its nuclear localization, promoting its degradation, or inactivating it through chemical modifications.
Repressors can indirectly repress transcription by recruiting histone modifiers deacetylases and methylases or nucleosome remodeling enzymes that affect the accessibility of the DNA. The elongation phase starts once assembly of the elongation complex has been completed, and progresses until a termination sequence is encountered. For example, the transcriptional activator Tat affects elongation rather than initiation during its regulation of HIV transcription. Other factors can also influence the stability and duration of the paused polymerase. Transcription termination has also emerged as an important area of transcriptional regulation.
Termination is coupled with the efficient recycling of polymerase. When transcription is arrested by the presence of a lesion in the transcribed strand of a gene, DNA repair proteins are recruited to the stalled RNA polymerase to initiate a process called transcription-coupled repair. TFIIH causes a conformational change in the polymerase, to expose the transcription bubble trapped inside, in order for the DNA repair enzymes to gain access to the lesion. Eukaryotic transcription is more complex than prokaryotic transcription.
For instance, in eukaryotes the genetic material DNA , and therefore transcription, is primarily localized to the nucleus, where it is separated from the cytoplasm in which translation occurs by the nuclear membrane. This allows for the temporal regulation of gene expression through the sequestration of the RNA in the nucleus, and allows for selective transport of mature RNAs to the cytoplasm.
Bacteria do not have a distinct nucleus that separates DNA from ribosome and mRNA is translated into protein as soon as it is transcribed. The coupling between the two processes provides an important mechanism for prokaryotic gene regulation. Transcription factors are one of the groups of proteins that read and interpret the genetic "blueprint" in the DNA.
They bind to the DNA and help initiate a program of increased or decreased gene transcription. As such, they are vital for many important cellular processes. Below are some of the important functions and biological roles transcription factors are involved in:.
Eukaryotic Transcription Factors
In eukaryotes , an important class of transcription factors called general transcription factors GTFs are necessary for transcription to occur. Other transcription factors differentially regulate the expression of various genes by binding to enhancer regions of DNA adjacent to regulated genes. These transcription factors are critical to making sure that genes are expressed in the right cell at the right time and in the right amount, depending on the changing requirements of the organism.
Many transcription factors in multicellular organisms are involved in development.
Navigation menu
The Hox transcription factor family, for example, is important for proper body pattern formation in organisms as diverse as fruit flies to humans. Cells can communicate with each other by releasing molecules that produce signaling cascades within another receptive cell. If the signal requires upregulation or downregulation of genes in the recipient cell, often transcription factors will be downstream in the signaling cascade. Estrogen is secreted by tissues such as the ovaries and placenta , crosses the cell membrane of the recipient cell, and is bound by the estrogen receptor in the cell's cytoplasm.
The estrogen receptor then goes to the cell's nucleus and binds to its DNA-binding sites , changing the transcriptional regulation of the associated genes. Not only do transcription factors act downstream of signaling cascades related to biological stimuli but they can also be downstream of signaling cascades involved in environmental stimuli.
Examples include heat shock factor HSF , which upregulates genes necessary for survival at higher temperatures, [27] hypoxia inducible factor HIF , which upregulates genes necessary for cell survival in low-oxygen environments, [28] and sterol regulatory element binding protein SREBP , which helps maintain proper lipid levels in the cell. Many transcription factors, especially some that are proto-oncogenes or tumor suppressors , help regulate the cell cycle and as such determine how large a cell will get and when it can divide into two daughter cells.
Transcription factors can also be used to alter gene expression in a host cell to promote pathogenesis. A well studied example of this are the transcription-activator like effectors TAL effectors secreted by Xanthomonas bacteria. When injected into plants, these proteins can enter the nucleus of the plant cell, bind plant promoter sequences, and activate transcription of plant genes that aid in bacterial infection.
It is common in biology for important processes to have multiple layers of regulation and control. This is also true with transcription factors: Not only do transcription factors control the rates of transcription to regulate the amounts of gene products RNA and protein available to the cell but transcription factors themselves are regulated often by other transcription factors.
Below is a brief synopsis of some of the ways that the activity of transcription factors can be regulated:. Transcription factors like all proteins are transcribed from a gene on a chromosome into RNA, and then the RNA is translated into protein. Any of these steps can be regulated to affect the production and thus activity of a transcription factor.
An implication of this is that transcription factors can regulate themselves. For example, in a negative feedback loop, the transcription factor acts as its own repressor: If the transcription factor protein binds the DNA of its own gene, it down-regulates the production of more of itself. This is one mechanism to maintain low levels of a transcription factor in a cell.
- PURIFICATION AND CLONING OF TRANSCRIPTION FACTORS.
- Building social Europe through the open method of coordination.
- Transcription factor - Wikipedia!
- Eukaryotic transcription factors.?
In eukaryotes , transcription factors like most proteins are transcribed in the nucleus but are then translated in the cell's cytoplasm. Many proteins that are active in the nucleus contain nuclear localization signals that direct them to the nucleus. But, for many transcription factors, this is a key point in their regulation. Transcription factors may be activated or deactivated through their signal-sensing domain by a number of mechanisms including:. DNA within nucleosomes is inaccessible to many transcription factors. Some transcription factors, so-called pioneering factors are still able to bind their DNA binding sites on the nucleosomal DNA.
For most other transcription factors, the nucleosome should be actively unwound by molecular motors such as chromatin remodelers. In many cases, a transcription factor needs to compete for binding to its DNA binding site with other transcription factors and histones or non-histone chromatin proteins. Most transcription factors do not work alone. Many large TF families form complex homotypic or heterotypic interactions through dimerization. This collection of transcription factors, in turn, recruit intermediary proteins such as cofactors that allow efficient recruitment of the preinitiation complex and RNA polymerase.
Thus, for a single transcription factor to initiate transcription, all of these other proteins must also be present, and the transcription factor must be in a state where it can bind to them if necessary. Cofactors are proteins that modulate the effects of transcription factors. Cofactors are interchangeable between specific gene promoters; the protein complex that occupies the promoter DNA and the amino acid sequence of the cofactor determine its spatial conformation.
Transcription factors are modular in structure and contain the following domains: TAD is domain of the transcription factor that binds other proteins such as transcription coregulators. The DNA sequence that a transcription factor binds to is called a transcription factor-binding site or response element.
Transcription factors interact with their binding sites using a combination of electrostatic of which hydrogen bonds are a special case and Van der Waals forces. Due to the nature of these chemical interactions, most transcription factors bind DNA in a sequence specific manner.
However, not all bases in the transcription factor-binding site may actually interact with the transcription factor.
